Prevalence and antibiotic susceptibility patterns of pseudomonas aeruginosa in urinary tract infections in a Tertiary care hospital, Central Kerala: A retrospective study over 4 years
Abstract
Background and Objective: Pseudomons aeruginosa (P.aeruginosa) is an important uropathogenthat has shown varied antibiotic susceptibility patterns. This study aims to findout the changing trends in th eprevalence and antibiotic susceptibility patterns ofurinary isolates of P.aeruginosa over four consecutive years.
Methodology: A retrospective, record based study was conducted on all culture and sensitivity (C/S) reports ofurine samples obtained in the microbiology lab in a tertiary care centre, Central Kerala (January 2014 -December 2017). The C/S reports which were positive for significant growth of P.aeruginosa were analyzed to findout its prevalence and antibiotic susceptibility patterns. Descriptive statistics was used for data analysis and the results were expressed in percentages.
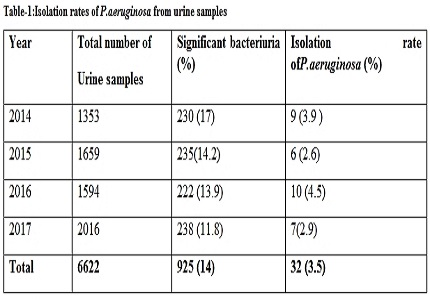
Result: Out of total 6622 urine samples received (14%) showed significant bacteriuria. P.aeruginosa was the third most common uropathogen isolated with an isolation rate of 3.5%. The antibiotic resistance observed were Gentamicin (53.1%), Amikacin (28%), Cefipime (28%), Ceftazidime(34.4%), Ciprofloxacin (43.7%), Norfloxacin (40.6%), Ofloxacin (40.6%), Piperacillin (37.5%), Piperacilli-Tazobactam (25%) and Imipenem (28%). The isolation rates of P.aeruginosa were 3.9%, 2.6%, 4.5 % and 2.9% in 2014, 2015, 2016 and 2017 respectively and overthe years it maintained its third position.The year wise analysis of antibiotic resistance showed fluctuating pattern except Amikacin, Cefipime and Fluoroquinoles which displayed a decreasingtrend. The reserve drugs like Piperacillin –tazobactam and Imipenem showed alarming drug resistance, although a hopeful reduction in the resistance was noted in 2017.
Conclusion: P.aeruginosa remains as a common uropathogen. Drug resistant strains are markedly high in our area. Antibiotic resistance of P.aeruginosado not show a consistent trend over years and vary from region to region. Soeach institution should have an antibiotic policy based on the local antibiogram which is to be renewed regularly. Instead of opting for higher antibioticseach time, strict implementation of restrictive and rotational antibiotic policies and adherence to the concept of ‘Reserve drugs” should be followed. This is the only modality to inhibit the emergence of resistance strains of all uropathogens especially opportunistic pathogens like P.aeruginosa.
Downloads
References
2. Singh VP, Mehta A. Bacteriological profile of urinary tract infections at a tertiary care hospital in Western Uttar Pradesh, India. Int J Res Med Sci. 2017 May;5(5):2126-9.DOI: http://dx.doi.org/10.18203/2320-6012.ijrms20171855
3. KL S, Rao G G, Kukkamalla A M. Prevalence Of Non-fermenters In Urinary Tract Infections In A Tertiary CareHospit.WebmedCentralMICROBIOLOGY.2011;2(1):WMC001464.doi: 10.9754/journal.wmc.2011.001464.
4. Shah DA, Wasim S, Abdullah FE. Antibiotic resistance pattern of Pseudomonas aeruginosa isolated from urine samples of Urinary Tract Infections patients in Karachi, Pakistan. Pak J Med Sci.2015;31(2):341-345.DOI:10.12669/pjms.312.6839.
5. Trivedi MK, BrantonA,Trivedi D, Nayak , Shettigar H et al. Antibiogram of Multidrug-Resistant Isolates of Pseudomonas aeruginosa after Biofield Treatment. J Infect Dis Ther.2015;3(5):244. doi:http://dx.doi.org/10.4172/2332-0877.1000244.
6. Somashekara SC, Deepalaxmi S, Jagannath N, Ramesh B, Laveesh MR, Govindadas D.Retrospective analysis of antibiotic resistance pattern to urinary pathogens in a Tertiary Care Hospital in South India.J BasicClin Pharm. 2014 Sep;5(4):105-8. doi: 10.4103/0976-0105.141948.
7. Biswas R, Rabbani R, Ahmed HS, Sarkar MAS, Zafrin N, Rahman MM. Antibiotic sensitivity pattern of urinary tract infection at a tertiary care hospital. Bangladesh Crit Care J. 2014;2(1):21-4.doi: http://dx.doi.org/10.3329/bccj.v2i1.19952.
8. Collee, J.G., Fraser AG, Marmion BP, Simmons A. Mackey & McCartney Practical Medical Microbiology. 14th ed. New Delhi, India: Elsevier; 2006.
9. Performance Standards for antimicrobial susceptibility testing; Clinical and laboratory standards institute. Twenty-Third Informational supplement. 2013; 33 (1):M100-S23.
10. Bency JAT, Priyanka R, Jose P. A study on the bacteriological profile of urinary tract infection in adults and their antibiotic sensitivity pattern in a tertiary care hospital in central Kerala, India. Int J Res Med Sci.2017;5(2):666-9. doi: http://dx.doi.org/10.18203/2320-6012.ijrms20170171.
11. Syed MA, Ramakrishna PJ, Shaniya K, Arya B , Shakir VP. Urinary Tract Infections – An overview on the Prevalence and the Anti-biogram of Gram Negative Uropathogens in A Tertiary Care Centre in North Kerala, India. J Clin Diag R.2012 September;6(7):1192-5.
12. Prakash D, Saxena RS. Distribution and Antimicrobial Susceptibility Pattern of Bacterial Pathogens Causing Urinary Tract Infection in Urban Community of Meerut City,India.ISRNMicrobiol.2013;749629. doi:10.1155/2013/749629.
13. Sangeeta Fattesingh Bhalavi, Vaishali Rahangdale, S.G. Joshi. Study of antibiotic resistance pattern to urinary pathogens in a tertiary care hospital in central India. Int J Sci Res.2017; 6(8): 428-429.
14. Shikha Jain, Geeta Walia, Rubina Malhotra. Prevalence and antimicrobial susceptibility patterns of ESBL producing gram negative bacilli in 200 cases of urinary tract infections. Int J pharm pharm Sci .2014; 6(10) : 210-211.
15. Oladeinde BH, Omoregie R, Olley M, Anunibe JA. Urinary tract infection in a rural community of Nigeria.N Am J Med Sci. 2011 Feb;3(2):75-7. doi: 10.4297/najms.2011.375.
16. Thomas SS, Sreenath K,Sebastian S. Characterization of the antibiotic profile of Pseudomonas aeruginosa isolates from a tertiary care center. Int J Res Med Sci. 2016;4:571-4 DOI: http://dx.doi.org/10.18203/2320-6012.ijrms20160317.
17. Juayang C, Lim T, BonifacioV , Lambot L, Millan SM, Sevilla N , Sy T, Villanueva PJ , Carmina P. Grajales and Gallega T . Five-Year Antimicrobial Susceptibility of Pseudomonas aeruginosa from a Local TertiaryHospital in Bacolod City, Philippines.Trop. Med. Infect. Dis. 2017;2(3):28 doi:10.3390/tropicalmed2030028 .
18. MohanasoundaramKM.The Antimicrobial Resistance Pattern in the Clinical Isolates of PseudomonasAeruginosa in a Tertiary Care Hospital;2008–2010 (A 3 Year Study).J Clin Diagn Res.2011 June;5(3):491-494.www.jcdr.net/articles/PDF/1375/2267.pdf
19. Olayinka B.O, Olonitola O.S, Olayinka A.T, Agada E.A. Antibiotic susceptibility pattern and multiple antibiotic resistance index of Pseudomonas aeruginosa urine isolates from a University teaching hospital. Afr. J. Clin. Exper.Microbiol.2004 May;5(2):198-202.
20. Jalal S, Wretlind B. Mechanisms of quinolone resistance in clinical strains of Pseudomonas aeruginosa.Microb Drug Resist. 1998 Winter;4(4):257-61. [PubMed]
21. Sharma N, Gupta AK, Walia G, Bakhshi R. A retrospective study of antimicrobial resistance pattern of Pseudomonas aeruginosa isolates from urine samples over last three years (2013-2015). Int J Basic Clin Pharmacol. 2016 ;5(4):1551-4. DOI: http://dx.doi.org/10.18203/2319-2003.ijbcp20162470.


 OAI - Open Archives Initiative
OAI - Open Archives Initiative


