In vitro methods of CLSI to detect Carbapenemases in Carbapenem-resistant Enterobacteriaceae
Sinha D.1, Shantala GB.2*, Ambica R.3, Kusuma CR.4
DOI: https://doi.org/10.17511/jopm.2020.i04.09
1 Deepa Sinha, Post-Graduate, Department of Microbiology, Bangalore Medical College and Research Institute, Bangalore, Karnataka, India.
2* Shantala G B., Associate Professor, Department of Microbiology, Bangalore Medical College and Research Institute, Bangalore, Karnataka, India.
3 Ambica R., Professor and Head, Department of Microbiology, Bangalore Medical College and Research Institute, Bangalore, Karnataka, India.
4 Kusuma CR., Lecturer, Department of Microbiology, Bangalore Medical College and Research Institute, Bangalore, Karnataka, India.
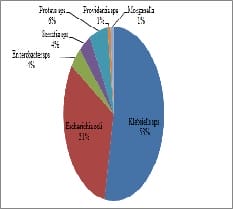
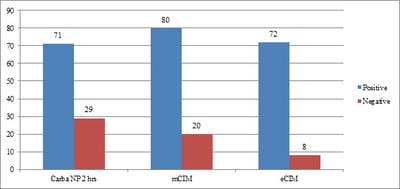
Aims: Detection of Carbapenemases production in Carbapenem Resistant Enterobacteriaceae(CRE) using CarbaNP and its comparison with the newer CLSI recommended modified Carbapenemases Inactivation Method(mCIM). Materials and Methods: 100 isolates of CRE were selected in a period of 3 months from June-August 2018 for the tests, based on MIC values of >= 2mg/ml given by vitek2compact systems. CarbaNP test was performed as per M100, CLSI 2018 guidelines. CarbaNP solutions A and B were prepared and Imipenem-Cilastatin powder was replaced for standard imipenem powder. The reading was taken at the end of 10 mins,30 mins, and 2 hours. mCIM was performed as per CLSI M100,2018. The presence of carbapenemase was indicated by an inhibition zone ≤15mm in diameter. eCIM was also performed in the same way, with 0.1M EDTA added in TSB broth. Descriptive analysis. Results: Among the 100 CRE isolates, Carba NP detected 71 CP-CRE while mCIM detected 80(80%) of the same. mCIM detected 8 more positives than the carbaNP test. 1 isolate tested positive CarbaNP but tested negative in mCIM. Conclusion: CarbaNP is a novel protocol recommended by CLSI for detecting CP-CREs and studies prove that it is a very good test in detecting KPC and OXA mediated resistance, while mCIM is said to detect all 5 genes i.e. KPC,NDM,OXA,IMP, and VIM. Carba NP is easy to perform and gives quick results once the reagents are prepared. mCIM on the contrary as a newly developed test detects more CP-CREs but takes more time for the interpretation.
Keywords: Carbapenemases, Enterobacteriaceae, CarbaNP, mCIM, CP-CRE
| Corresponding Author | How to Cite this Article | To Browse |
|---|---|---|
| , Associate Professor, Department of Microbiology, Bangalore Medical College and Research Institute, Bangalore, Karnataka, India. Email: |
Sinha D, Shantala GB, Ambica R, Kusuma CR. In vitro methods of CLSI to detect Carbapenemases in Carbapenem-resistant Enterobacteriaceae. Trop J Pathol Microbiol. 2020;6(4):324-328. Available From https://pathology.medresearch.in/index.php/jopm/article/view/457 |


 ©
©