Antibiogram of Klebsiella pneumoniae isolated from various clinical samples of hospitalized patients in a tertiary care hospital of North India
Kaur Gill M.1*, Kaur Gill A.2, Khanna A.3
DOI: https://doi.org/10.17511/jopm.2019.i08.01
1* Manmeet Kaur Gill, Associate Professor, Department of Microbiology, Sri Guru Ram Das Institute of Medical Sciences and Research, Amritsar, Punjab, India.
2 Anureet Kaur Gill, BDS Final Year Student, , Sri Guru Ram Das Institute of Dental Science and Research, Amritsar, Punjab, India.
3 Ashish Khanna, Professor, Department of Microbiology, Sri Guru Ram Das Institute of Medical Sciences and Research, Amritsar, Punjab, India.
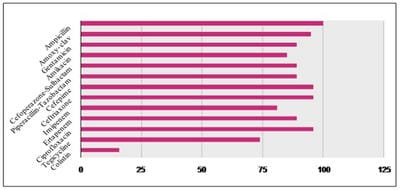
Background: Klebsiella pneumoniae is an important cause of nosocomial and community acquired infections worldwide. It exhibits high antibiotic resistance due to production of Extended Spectrum Beta Lactamases (ESBLs) and Carbapenamases. The aim of present study was to know its resistance pattern of Klebsiella pneumoniae so as to help local physicians choose appropriate antibiotics for effective infection control. Materials and Methods: It was a prospective study carried out from January 2019 to July 2019 in the department of Microbiology of a tertiary care hospital in North India. The study comprised of a total of 194 non-repeat isolates obtained from various clinical samples received in Microbiology lab for culture & sensitivity testing. All isolates were processed to determine their antimicrobial sensitivity profile. For data analysis SPSS software, version 17.0 and MS excel 2007 were used. Results: Out of total 2155 (22.43%) positive cultures 194 (9%) isolates were that of Klebsiella pneumoniae Most of the isolates obtained were multi-drug resistant, ESBL and Carbapenamase producers. 100% isolates showed resistance to Ampicillin.Conclusion: Since the frequency of multiple drug resistance among Klebsiella pneumoniae is alarmingly high, therefore periodic monitoring of antimicrobial susceptibility profile of these agents is much needed. This will help physician in selection of appropriate chemotherapy and thus help in effective management of the infections and better patient care.
Keywords: Klebsiella pneumoniae, Clinical samples, Antimicrobial resistance profile
| Corresponding Author | How to Cite this Article | To Browse |
|---|---|---|
| , Associate Professor, Department of Microbiology, Sri Guru Ram Das Institute of Medical Sciences and Research, Amritsar, Punjab, India. Email: |
Gill MK, Gill AK, Khanna A. Antibiogram of Klebsiella pneumoniae isolated from various clinical samples of hospitalized patients in a tertiary care hospital of North India. Trop J Pathol Microbiol. 2019;5(8):512-516. Available From https://pathology.medresearch.in/index.php/jopm/article/view/297 |


 ©
©