Utility of nested PCR in diagnosing extra-pulmonary Tuberculosis
Kaur Gill M.1*, Khanna A.2, Sharma S.3
DOI: https://doi.org/10.17511/jopm.2019.i07.05
1* Manmeet Kaur Gill, Associate Professor, Department of Microbiology, Sri Guru Ram Das Institute of Medical Sciences and Research, Amritsar, Punjab, India.
2 Ashish Khanna, Professor, Department of Microbiology, Sri Guru Ram Das Institute of Medical Sciences and Research, Amritsar, Punjab, India.
3 Sarbjeet Sharma, Professor and Head, Department of Microbiology, Sri Guru Ram Das Institute of Medical Sciences and Research, Amritsar, Punjab, India.
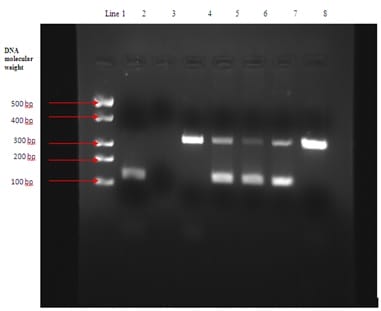
Background: Tuberculosis has always been a burden for developing countries such as India. Conventional techniques (smear microscopy and culture) used for diagnosing this highly contagious disease have a lower sensitivity in comparison to the advanced diagnostic techniques. Polymerase chain reaction has emerged as a better alternative with higher specificity and sensitivity. This study was therefore designed for evaluating the efficacy of IS6110 sequence based nested polymerase chain reaction (nPCR) in detecting Mycobacterium tuberculosis (MTB) in extra-pulmonary clinical samples. Material and Methods: The study comprised of 195 samples of extra-pulmonary origin obtained from the patients with history suggestive of tuberculosis. The samples were immediately processed and subjected to Ziehl-Neelsan (ZN) staining for presence of acid fast bacilli (AFB), inoculation on Lowenstein-Jensen (LJ) medium for culture and nested PCR using specific primers that target the 123bp fragment of IS6110 of the MTB- complex. Results: It was observed that 72/195 (36.92%) samples were diagnosed positive for TB using PCR whereas only 29/195 (14.87%) were positive by smear and 32/195 (16.41%) by culture on LJ. Total positive samples by conventional methods (either smear or culture) were 35/195 (17.94%). These results clearly indicated that PCR had higher sensitivity over conventional methods with a significant difference in p value (p<0.05). Conclusions : It was concluded from the study that nPCR method targeting IS6110 gene sequence is more efficient, rapid, sensitive, and specific method for detecting Mycobacterium tuberculosis genetic material in extra-pulmonary tuberculosis specimens of patients with suspected history of TB in contrast to the available conventional methods.
Keywords: Mycobacterium tuberculosis, Conventional methods, Nested polymerase chain reaction, IS6110 sequence
| Corresponding Author | How to Cite this Article | To Browse |
|---|---|---|
| , Associate Professor, Department of Microbiology, Sri Guru Ram Das Institute of Medical Sciences and Research, Amritsar, Punjab, India. Email: |
Kaur Gill M, Khanna A, Sharma S. Utility of nested PCR in diagnosing extra-pulmonary Tuberculosis. Trop J Pathol Microbiol. 2019;5(7):443-448. Available From https://pathology.medresearch.in/index.php/jopm/article/view/286 |


 ©
©